CherryPicker parameterisation algorithm class. More...
#include <indigox/algorithm/cherrypicker.hpp>
Public Types | |
| enum | Settings : uint8_t |
| User controllable settings for the CherryPicker algorithm. More... | |
Public Member Functions | |
| CherryPicker (Forcefield &ff) | |
| Constructor. More... | |
| bool | AddAthenaeum (Athenaeum &library) |
| Add an Athenaeum for parameterisation purposes. More... | |
| void | DefaultSettings () |
| Set the default values for all of the settings. More... | |
| bool | GetBool (Settings param) |
| Get the current state of a boolean setting. More... | |
| Forcefield | GetForcefield () |
| Get the assigned forcefield. More... | |
| int32_t | GetInt (Settings param) |
| Get the current value of an integer setting. More... | |
| int32_t | NumAthenaeums () |
| The number of Athenaeums in the list. More... | |
| ParamMolecule | ParameteriseMolecule (Molecule &mol) |
| Apply the CherryPicker algorithm to a molecule. More... | |
| bool | RemoveAthenaeum (Athenaeum &library) |
| Remove an Athenaeum from the list. More... | |
| void | SetBool (Settings param) |
| Set the state of a boolean setting to true. More... | |
| void | SetInt (Settings param, int32_t value) |
| Set the value of an integer setting. More... | |
| void | UnsetBool (Settings param) |
| Set the state of a boolean setting to false. More... | |
Detailed Description
CherryPicker parameterisation algorithm class.
The CherryPicker algorithm is a parameterisation algorithm for molecular dynamics simulations. It parameterises novel molecules by identifying fragments of previously parameterised molecules which match portions of the novel molecule, then applying the parameters found to the novel molecule. See the tutorial for a brief tutorial on how to parameterise something using the CherryPicker algorithm.
There are three data classes and one graph class primarily associated with CherryPicker. The Athenaeum class acts as a library of previously parameterised molecules which can be used to parameterise a target molecule. Within an Athenaeum, each Molecule is mapped to a vector of Fragment instances. The Fragment class represents a fragment of a source molecule. It is the fragments which are checked for matching portions of the target molecule. This checking is performed using the CondensedMolecularGraph (CMG) of the target molecule and the fragment. Accounting of the matches found is performed by the ParamMolecule class and its components.
To parameterise a novel molecule using CherryPicker, first an Athenaeum must be created (see the Athenaeum documentation). For a given Athenaeum, the process of parameterisation is as follows. Fragments within the Athenaeum are iterated through in an arbitary order. The CMG of each fragment is checked against the CMG of the target molecule for subgraph isomorphism. All subgraph isomorphisms between the two graphs are enumerated, giving a series of one-to-one mappings between the vertices of each graph. These mappings are expanded to give one-to-one mappings between the atoms of the fragment's source molecule and the target molecule. The use of all possible mappings between the condensed atoms of each vertex is available through the Settings. From these mappings, the atoms, bonds, angles and dihedrals that match one another from the fragment source molecule and the target molecule are known. Parameters assigned to these components in the fragment source molecule are accounted for by adding them to the corresponding component within the ParamMolecule created from the target molecule. Once all fragments within an Athenaeum have been checked, any component within the ParamMolecule which has been mapped by at least one component from a fragment applies a parameter back to the corresponding component within the target molecule. In the case of discrete parameters, such as bond types, the parameter applied is the mode of all mapped parameters. In the case of continuous parameters, such as atomic partial charges, the parameter applied is the mean of all mapped parameters. If an Athenaeum is marked as being self-consistent, it is at the point of applying these parameters that the self-consistency is checked. For discrete parameters, self-consistent is defined as having only one mapped type on the ParamMolecule component. For continuous parameters, it is defined as having a difference of no greater than 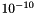 between the largest and smallest mapped values.
between the largest and smallest mapped values.
CherryPicker is designed to support multiple Athenaeums. This is handled in a first in-first out (FIFO). That is, Athenaeums are iterated through in the order in which they were assigned to the CherryPicker instance. When multiple Athenaeums are provided, parameterisation occurs as above for each Athenaeum. Additionally, once all fragments within an Athenaeum have been checked and parameters applied, any components which had a parameter applied is removed from the set of parameterisable components so that no further Athenaeums will be able to parameterise it. The reasoning for this FIFO methadology is so that portions of a molecule can be parameterised by a smaller set of well tested fragments. For example, a protein with a few unnatural or unique residues can initially be parameterised with fragments matching the natural amino acids, thus covering the majority of the molecule quickly, before a more shotgun approach to parameterising the remaining portions is utilised.
An exception to the rule of not further parameterising a component which has been parameterised by a previous Athenaeum is when any excess charge is to redistributed across the molecule. In this case, all atoms which were not parameterised by a self-consistent Athenaeum are available to have extra charge added.
The primary working component of the algorithm is subgraph isomorphism. Whenever a fragment is tested to check if it matches the test molecule, the CMG of the fragment is tested for subgraph isomorphism with the CMG of the target molecule.
There are two kinds of settings, boolean and integer values. Attempting to modify a setting through the incorrect access type (line 4 of the above code snippet) will result in an error being thrown. See the Settings documentation for details.
The ParameteriseMolecule() method applies the CherryPicker algorithm to the provided molecule. Though it returns a ParamMolecule, it is safe to ignore this as the parameters discovered are applied directly to the provided molecule. If you desire to modify the parameters discovered, the returned ParamMolecule provides all of the information relating to what parameters were found for each component of the molecule. See the Molecule class for serialisation options.
Member Enumeration Documentation
◆ Settings
|
strong |
User controllable settings for the CherryPicker algorithm.
Settings are maintained on a per-instance basis. That is multiple different CherryPicker instances can have different settings. There are two types of settings, boolean and integer. Boolean settings are either on or off, and are manipulated through the SetBool() and UnsetBool() methods. These settings appear before the BoolCount marker. Integer settings require an integer value and are manipulated through the SetInt() method. These settings appear between the BoolCount and IntCount markers. Default values are detailed in the DefaultSettings() method.
| Enumerator | |
|---|---|
| VertexElement | When performing subgraph isomorphism testing, two vertices will only match if the element of their associated atoms are the same. |
| VertexFormalCharge | When performing subgraph isomorphism testing, two vertices will only match if the formal charge of their associated atoms are the same. |
| VertexCondensed | When performing subgraph isomorphism testing, two vertices will only match if the counts per type of condensed vertices are the same. |
| VertexCyclic | When performing subgraph isomorphism testing, two vertices will only match if they are both contained within a cycle. To be regarded as in a cycle, the size of the smallest cycle a vertex is in must be less than or equal to the ??? setting.
|
| VertexCyclicSize | When performing subgraph isomorphism testing, two vertices will only match if the smallest cycle they are both contained in have the same size. |
| VertexStereochemistry | When performing subgraph isomorphism testing, two vertices will only match if the assocaited atoms have the same stereochemistry. |
| VertexAromaticity | When performing subgraph isomorphism testing, two vertices will only match if the assocaited atoms have the same aromaaticity. |
| VertexDegree | When performing subgraph isomorphism testing, two vertices will only match if the assocaited atoms have the same number of bonds. |
| EdgeBondOrder | When performing subgraph isomorphism testing, two edges will only match if the bond order of their associated bonds are the same. |
| EdgeStereochemistry | When performing subgraph isomorphism testing, two edges will only match if their assocaited bonds have the same stereochemistry. |
| EdgeCyclic | When performing subgraph isomorphism testing, two edges will only match if they are both contained within a cycle. To be regarded as in a cycle, the size of the smallest cycle a edge is in must be less than or equal to the ??? setting.
|
| EdgeCyclicSize | When performing subgraph isomorphism testing, two edges will only match if the smallest cycle they are both contained in have the same size. |
| EdgeDegree | When performing subgraph isomorphism testing, two edges will only match if the atoms of the associated bond have the same number of bonds. That is, the atom of each bond with the lowest number of bonds must have the same number of bonds, and the same for the atom with the highest number of bonds. |
| AllowDanglingBonds | When applying parameters to a bond, allow for parameteristion across the overlap region. If one atom of a bond is in the fragment region and the other is in the overlap, setting this allows the matching bond to be parameterised by the bond between the fragment and overlap regions. |
| AllowDanglingAngles | When applying parameters to an angle, allow for parameteristion across the overlap region. If two atoms of a angle are in the fragment region and the other is in the overlap, setting this allows the matching angle to be parameterised by the angle between the fragment and overlap regions. |
| AllowDanglingDihedrals | When applying parameters to a dihedral, allow for parameterisation across the overlap region. If two or three atoms of a dihedral are in the fragment region and the others are in the overlap region, setting this allows the matching dihedral to be parameterised by the dihedral between the fragment and overlap regions. This only applies to dihedrals where the atoms that are in the fragment and not the overlap are adjacent to one another. |
| ParameteriseFromAllPermutations | When applying parameters iterate through all permutations of contracted vertex mappings. On any given vertex match, contracted vertices of the same type can be mapped to one another in all possible permutations. This setting causes all such permutations to be used for applying parameters. In general, this is not required as contracted vertices are expected to have the same parameters.
|
| BoolCount | Marks the end of the boolean settings. As there is no external use for this value, it is not exposed to Python. |
| MinimumFragmentSize | The minimum size of fragments to check for matching purposes. Any fragment within an athenaeum which has a size smaller than this will be skipped. A negative value means all fragments will be tested. |
| MaximumFragmentSize | The maximum size of fragments to check for matching purposes. Any fragment in an athenaeum which has a size larger than this will be skipped. A negative value means all fragments will be tested. |
| IntCount | Marks the end of the integer settings. As there is no external use for this value, it is not exposed to Python. |
Constructor & Destructor Documentation
◆ CherryPicker()
| CherryPicker | ( | Forcefield & | ff | ) |
Constructor.
- Parameters
-
ff defines what forcefield will be used for parameterisation.
Member Function Documentation
◆ AddAthenaeum()
| bool AddAthenaeum | ( | Athenaeum & | library | ) |
Add an Athenaeum for parameterisation purposes.
Adds an Athenaeum in a FIFO manner. No check is performed to see if library has previously been added, so multiple instances of the same Athenaeum is possible. Only check performed is that the Athenaeum's forcefield matches the CherryPicker forcefield.
- Parameters
-
library the Athenaeum to add.
- Returns
- if
librarywas successfully added.
◆ DefaultSettings()
| void DefaultSettings | ( | ) |
Set the default values for all of the settings.
The following boolean values are default set to true: VertexElement, VertexFormalCharge, VertexCondensed, VertexDegree, EdgeBondOrder, EdgeDegree, AllowDanglingBonds, AllowDanglingAngles, and AllowDanglingDihedrals. All other boolean values default to false. The default MinimumFragmentSize is 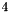 and the default MaximumFragmentSize is
and the default MaximumFragmentSize is 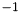 .
.
◆ GetBool()
| bool GetBool | ( | Settings | param | ) |
Get the current state of a boolean setting.
- Parameters
-
param The setting to get the state of.
- Returns
- the current state of the
paramsetting.
- Exceptions
-
std::runtime_error if paramis not a valid boolean setting.
◆ GetForcefield()
|
inline |
Get the assigned forcefield.
- Returns
- the assigned Forcefield.
◆ GetInt()
| int32_t GetInt | ( | Settings | param | ) |
Get the current value of an integer setting.
- Parameters
-
param The setting to get the value of.
- Returns
- the current value of the
paramsetting.
- Exceptions
-
std::runtime_error if paramis not a valid integer setting.
◆ NumAthenaeums()
|
inline |
The number of Athenaeums in the list.
- Returns
- the number of Athenaeums.
◆ ParameteriseMolecule()
| ParamMolecule ParameteriseMolecule | ( | Molecule & | mol | ) |
Apply the CherryPicker algorithm to a molecule.
Fill in the details here.
- Parameters
-
mol the molecule to parameterise.
- Returns
- a ParamMolecule for all the matched parameters.
- Exceptions
-
std::runtime_error if the list of Athenaeums is empty or molis not connected.
◆ RemoveAthenaeum()
| bool RemoveAthenaeum | ( | Athenaeum & | library | ) |
◆ SetBool()
| void SetBool | ( | Settings | param | ) |
Set the state of a boolean setting to true.
- Parameters
-
param The setting to set the state of.
- Exceptions
-
std::runtime_error if paramis not a valid boolean setting.
◆ SetInt()
| void SetInt | ( | Settings | param, |
| int32_t | value | ||
| ) |
Set the value of an integer setting.
- Parameters
-
param The setting to set the value of. value The value to set paramto.
- Exceptions
-
std::runtime_error if paramis not a valid integer setting.
◆ UnsetBool()
| void UnsetBool | ( | Settings | param | ) |
Set the state of a boolean setting to false.
- Parameters
-
param The setting to set the state of.
- Exceptions
-
std::runtime_error if paramis not a valid boolean setting.
The documentation for this class was generated from the following file:
- include/indigox/algorithm/cherrypicker.hpp
