Table of Contents
This page provides tutorials on how to use some of the classes and algorithms within the indigoX package. All these tutorials assume that you are using the provided Python bindings. The examples shown can all be run within the Python interpreter, with the proviso that the following two lines are included:
How to use CherryPicker
The CherryPicker algorithm is a parameterisation algorithm for molecular dynamics simulations. It parameterises novel molecules by identifying fragments of previously parameterised molecules which match portions of the novel molecule, then applying the parameters found to the novel molecule. This tutorial assumes that you are familar with molecule dynamics and parameterisation. A full working version of the code presented here can be found in the examples directory. For more in depth information about the algorithm, consult the CherryPicker documentation.
Overview
There are two main parts to using CherryPicker, producing Athenaeums to store fragments of previously parameterised molecules, and using these Athenaeums to apply the algorithm to parameterising a novel molecule. Fragments to store can either be automatically generated by the Athenaeum, or generated as defined in a fragment file. One of main reasons for the use of Athenaeums is to enable one to generate an Athenaeum from some data set once and then save it to disk to enable reuse at a later date. Finally, the CherryPicker algorithm has a number of user controllable settings to dictate how the algorithm should behave. Use of all these aspects will be discussed, but first...
First Steps
Before beginning, ensure that you have all the required files. Navigate to the examples/CherryPicker directory. This directory should contain three items: a python script (cherrypicker_example.py) containing the code presented here, and two subdirectories, SourceMolecules and TestMolecules. The SourceMolecules directory contains the previously parameterised molecules that we will be using for generating Athenaeums. There are twenty-two such molecules in total. Each molecule should have four files: a coordinate file (name_ua.pdb), a topology file (name.top), a fragment definition file (name.frag), and a data file (name.ixd). Within the TestMolecules subdirectory, there are six files: a coordinate file (name.pdb) and a data file (name.ixd) for each of three molecules.
The first step on any parameterisation undertaking is to obtain a forcefield:
Here, we use the included GROMOS 54A7 forcefield generator. Other methods for obtaining a forcefield are detailed in the forcefield documentation. The forcefield chosen should be the same as the forcefield your previously parameterised molecules were parameterised with. This forcefield is provided when creating both an Athenaeum and a CherryPicker instance. When adding an Athenaeum to CherryPicker, it will only be successfully added if the forcefields used to create both the Athenaeum and CherryPicker instance match. Next, we move on to discussing Athenaeum generation.
Athenaeum generation
As previously mentioned, there are two ways to add fragments to an Athenaeum, either by automatically generating all possible fragments of a given molecule, or by adding a fragment defined within a fragment definition file. There are also two types of Athenaeum, self-consistent and not self-consistent. A self-consistent Athenaeum is where if multiple fragments from the Athenaeum match to the same portion of a target molecule, all areas of overlapping matching will have the same parameters in each of the fragments. Generally speaking, you would not want to automatically generate fragments for a self-consistent Athenaeum.
Creating an Athenaeum is simple, just pass a forcefield and, optionally, an overlap length to the Athenaeum constructor.
Here, we create two. The first, man_ath, is a self-consitent Athenaeum to which we will add all the defined fragments from the SourceMolecules subdirectory. As we are not automatically generating fragments using this Athenaeum, the overlap length is irrelevant. The second, auto_ath, is created with an overlap length of one and will be used to generate all possible fragments from each of the molecules within the SourceMolecules subdirectory. As some of our source molecules are larger than the default upper size limit for a molecule to be able to be automatically fragmented, we must change the setting.
- Note
- Care should be taken with changing the upper size limit. The fragment generation algorithm technically scales as
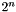 , where
, where 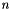 is the number of vertices in the condensed molecular graph. Practically, the scaling is not as bad as this due to the nature of chemical graphs. Nevertheless, with too large a molecule, the computational time required can become unrealistic.
is the number of vertices in the condensed molecular graph. Practically, the scaling is not as bad as this due to the nature of chemical graphs. Nevertheless, with too large a molecule, the computational time required can become unrealistic.
Once the Athenaeums have been prepared, all that remains is generate the fragments.
- Note
- Execution of this block of code can take a substainial time, and use a large amount of memory. Locally, this equated to an execution time of approximately 400 seconds and a maximum memory usage of around 3.5 GB.
Lets go through this line by line. The first line locates all the fragment definition files within the SourceMolecules directory, so the loop will be executed 22 times, once for each source molecule. The next line loads the fragment file. The LoadFragmentFile() function returns the molecule which the fragments are from, and the list of all fragments defined within the file. The forcefield is provided as internally, the LoadParameterisedMolecule() function is called to load the molecule. Line three, the inner loop, adds all of the fragments which were defined in the fragment file into the first athenaeum. Here we do not check the return value of the AddFragment() method, but it general it should be checked to ensure that the fragment was correctly added. For the fragment to be successfully added, its source molecule must be parameterised and the forcefield used for parameterisation must match the forcefield used to create the Athenaeum. The final line adds all possible fragments from the loaded molecule to the second Athenaeum. See the AddAllFragments() method documentation for a description of the algorithm used.
Finally, we save the Athenaeums to disk. The files are saved in a portable binary format, meaning they should work across different computer systems.
For further Athenaeum information, consult the Athenaeum documentation.
Applying CherryPicker algorithm
To discuss the application of the CherryPicker algorithm we assume a clean slate. That is, that the previous Athenaeum generation has been run, but the saved files need to be reloaded. As such, the first step is to load the Athenaeums we wish to parameterise from.
Next, we must create an CherryPicker instance. To ensure that the forcefield we create the instance with matches that of the Athenaeums we are going to add, we first retrieve the forcefield from one of the Athenaeums.
Now it is time to add the Athenaeums to the CherryPicker instance. For this example, we have two Athenaeums, a small self-consistent one and a much larger automatically generated one. As we are more confident in the parameters within the small Athenaeum, and that CherryPicker iterates through Athenaeums in a first in-first out manner, the small Athenaeum should be added first.
And change some of the default CherryPicker settings.
With this setup performed, we are ready to parameterise our test molecules. As there are three test molecules, we loop over them, applying the parameteristion and saving the resultant parameters to a GROMACS ITP file.
By providing the returned parameterised molecule to the save function, the output ITP file will contain additional statistics related to the parameterisation.
